Platform Technologies of OncoTherapy Science, Inc.
To develop new anti-cancer medicines with a minimal risk of adverse events, it is important to identify suitable target genes. We, therefore, applied laser microbeam microdissection (LMM) that obtains a high pure population of cancer cells from cancer tissues and genome-wide cDNA microarray. Together with these technologies that provide comprehensive gene expression profile analyses and gene function analyses as fundamental technologies, we have identified suitable target genes.
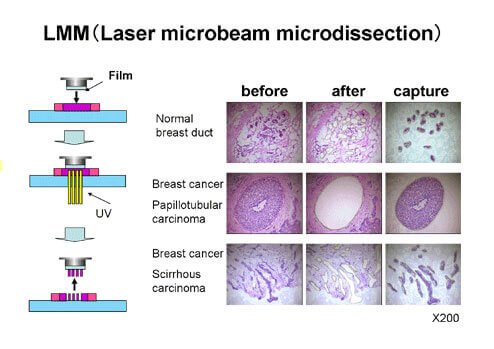
Laser Micro-beam Microdissection (LMM)
Human cancer tissues consist of both cancer cells and non-cancerous cells including stromal cells and normal cells. Even if gene expression analysis of such cancer tissues as samples is carried out, it is not possible to obtain gene expression profiles of pure cancer cells. LMM enables us to obtain an almost 100% pure population of cancer cells from clinical cancer tissues. We are using this technology and analyzing genes specifically expressed in cancer cells by using captured cancer cells as samples.
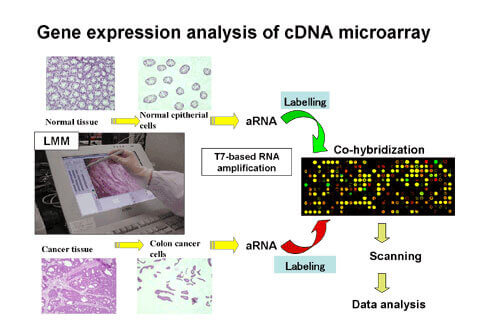
cDNA Microarray
We established a unique cDNA microarray system with high sensitivity and specificity to obtain expression profiles of almost all human genes. We amplified a single copy part of cDNA 3′ (200 and 1000 base pairs) using a primer set specific to the cDNA sequences to prepare spotting target genes. A gene-specific signal with high sensitivity may be obtained by spotting these PCR products in a high density on a special glass slide. The combination of LMM and our cDNA microarray system enables us to provide accurate, high quality and reliable expression profile data. In addition, gene expression analysis in normal organs is performed using our cDNA microarray system, and we are carrying out analysis of expression patterns in normal organs.
Identification of Target Genes
We identified gene groups that were highly expressed in cancer cells but not in any normal or vital organs that are essential for life by gene expression profile using LMM and our cDNA microarray system. These genes were categorized into two groups. One is that genes over-expressed in cancer cells, and the other is that the upregulation of genes is required for carcinogenesis. We then applied the RNA interference technology to examine gene functions whether genes selected by gene expression information are essential for the growth or survival of cancer cells. We focus on the genes which inhibit the growth or survival of cancer by RNA interference for drug development research.
Following is the three criteria for OTS’s target identification with our fundamental technology.
- ・Over-expressed in cancer cells
- ・Absent or hardly detectable in normal organs
- ・Essential for the growth or survival of cancer cells.
